Einleitung
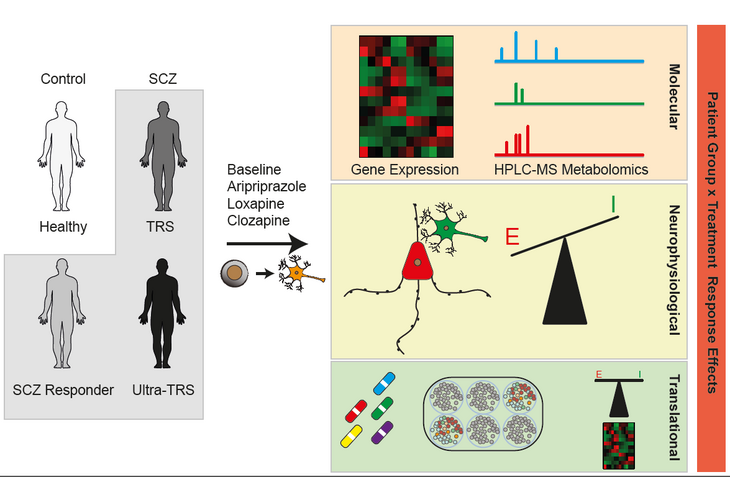
Up to one fourth of all SCZ patients will eventually convert to a treatment resistant state and typically receive clozapine as last resort treatment. At present, the biological mechanisms underlying the emergence of treatment resistant schizophrenia (TRS) remain unclear, but multiple lines of evidence suggest a strong genetic component. In order to dissect this molecular genetic basis of TRS, the Ziller group has generated induced pluripotent stem cells (iPSCs) from a large cohort of healthy individuals, first line responder and TR SCZ patients. These iPSCs were differentiated into different types of cortical neurons and subsequently investigated under baseline and treatment in vivo conditions, administering aripiprazole, loxapin and clozapine. Subsequently, treated and untreated neuronal cultures were subjected to deep molecular (transcriptomics, metabolomics) and electrophysiological (multi-electrode-arrays, matching patient EEG in collaboration with the LMU Munich) characterization. These datasets are all readily available. It is the central goal of the overarching project to pinpoint genes, metabolites and electrophysiological parameters associated with treatment resistance and treatment response in vivo for subsequent follow up studies.
Fragestellung
Within this complex matrix of questions, the focus of offered PhD thesis is on the analysis of the electrophysiological data from iPSC derived neurons and (matching) patients. In particular, it is the central goal of this thesis to identify metabolites and metabolic pathways differentially regulated between patient groups under baseline and treatment conditions. The student will specifically test the long standing hypothesis in the field of altered neurotransmitter concentrations and energy metabolism in the respective cells.
Bearbeitung:
The PhD student will initially receive training datasets and familiarize her/himself with computational analysis methods for multi-electrode-array (MEA) and EEG data. Following successful analysis and quality control of these training data, the student will receive the relevant pre-processed data from this study for analysis.
Methoden
- Computational analysis of high dimensional data in R
- Quality control of MEA data
- Basic and complex analysis techniques for MEA/EEG data using generalized linear models, meaTools, spectrum analysis (e.g. FFT)
- Identification and control of confounding factors in complex datasets using surrogate variable analysis and combat
- Correlation analyses, dimensionality reduction techniques
Arbeitsprogramm
1. Training in R, MEA analysis and analysis of example datasets. Introduction to github.
2. Quality control and filtering of MEA data from treated and untreated SCZ patients and controls. Identification of confounders, evaluation of correction methods for confounders
3. Identification of relevant measurement variables in MEA and EEG data
4. Differential activity using linear mixed models and network activity analysis
5. Differential EEG analysis of TRS and non-TRS patients using linear models
6. Translational analysis, correlating in vivo EEG and in vitro MEA result
7. Documentation of analyses and results through online code repositories and Jupiter notebooks.