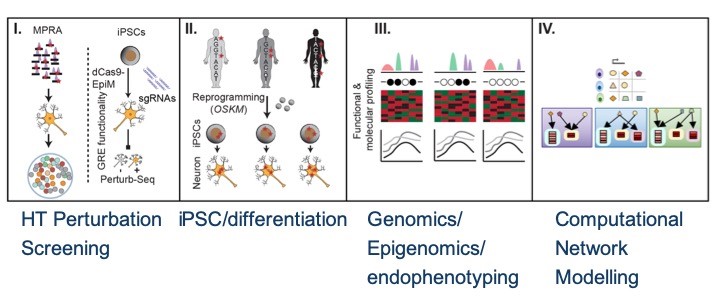
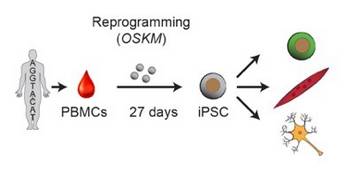
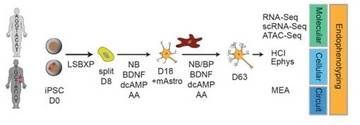
Beyond GWAS, translating genetic associations underlying psychiatric disorders into neurobiological insights using functional genomics and induced pluripotent stem cell models
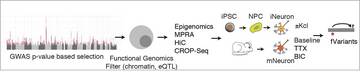
It is the mission of my group to develop novel paradigms to understand the genetic basis of complex genetic and in particular psychiatric diseases such as schizophrenia and bipolar disorder. To implement this goal, the lab for functional genomics in psychiatry pursues a combined experimental and computational strategy to deconstruct the polygenic risk architecture underlying complex diseases with the goal to decode the molecular mechanisms that contribute to disease onset and progression and treatment resistance.
Moreover, it is the central goal of my lab to rapidly operationalize these insights to optimize patient treatment, obtain insights into the molecular cause of treatment resistance and identify new drug targets.